Fumarate Hydratase
(Fumarase)
The metabolic role of fumarase:
Fumarate hydratase catalyzes the seventh reaction of the tricarboxylic acid (TCA) cycle, or Krebs cycle, in which acetyl-CoA from glycolysis produces CO2, reduced electron carriers (NADH and FADH2) and a small amount of ATP.
In this reversible reaction, the carbon-carbon double bond of fumarate is hydrated to produce L–malate with a free energy change of -3.8 kJ/mol:

![]()
![]()


Fumarate
L-Malate
ΔGo′ = -3.8 kJ/mol
Figure 1.
The reversible conversion of fumarate to malate, catalyzed by fumarate
hydratase.
About the enzyme:
Fumarate hydratase exists in two isoforms: a cystolic form and mitochondrial form. The mitochondrial form has an extended N-terminus and is responsible for the reaction described above, whereas the cytosolic form metabolizes fumarate, a by-product of the urea cycle.
In humans, fumarate hydratase is coded for by the gene FH, which has been mapped to chromosome 1 at 1q42.1 (Despoisses et al. 1984) and consists of 22,149 basepairs. 510 amino acids comprise the gene product (molecular weight 54,637 Da), which is highly conserved across species (Weaver et al. 1995).
Fumarate is a tetrameric enzyme, composed of four identical subunits of 50 kDa each and the residues from three of the chains form the active site (site A) of the enzyme (Weaver and Banaszak1996). In addition to binding the substrates, the active site binds their analogs D-malate and oxaloacetate, and the competitive inhibitor glycine. An additional binding site (site B), which has a lower affinity for binding ligands, binds the substrates and their analogs as well.
Known inhibitors of
the enzyme include (Massey 1952):
-
competitive
inhibitors: D-malic, trans-aconitic,
citric, mesaconic, maleic, adipic, glutaric, succinic, malonic, D-tartaric
acids, glycine
-
Non-competitive
inhibitors: thiocyanate, acetylenedicarboxylic acid
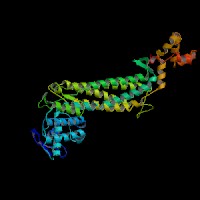
Figure 2.
ModBase 3D structure of fumarate hydratase in humans, viewed from (A) the top, and
(B) the side. Source: http://modbase.compbio.ucsf.edu/modbase-cgi-new/search_form.cgi.
A homozygous mutation of FH leads to fumarate deficiency (causing delayed development, diminished muscle tone and degenerative brain disease), whereas a heterozygous mutation of FH causes multiple cutaneous and uterine leiomyomata (MCUL, characterized by benign smooth muscle tumors of the skin and uterus) and renal cell cancer (Alam et al. 2003).
Resources
Alam NA, Rowan AJ, Wortham NC, Pollard PJ, Mitchell M,
Tyrer JP, Barclay E, Calonje E, Manek S, Adams SJ, Bowers PW, Burrows NP,
Charles-Holmes R, Cook LJ, Daly BM, Ford GP, Fuller LC, Hadfield-Jones SE,
Hardwick N, Highet AS, Keefe M, MacDonald-Hull SP, Potts EDA, Crone M,
Wilkinson S, Camacho-Martinez F, Jablonska S, Ratnavel R, MacDonald A, Mann RJ,
Grice K, Guillet G, Lewis-Jones MS, McGrath H, Seukeran DC, Morrison PJ,
Fleming S, Rahman S, Kelsell D, Leigh I, Olpin S, Tomlinson IPM. (2003) Genetic and functional analyses of FH
mutations in multiple cutaneous and uterine leiomyomatosis, hereditary
leiomyomatosis and renal cancer, and fumarate hydratase deficiency. Human
Molecular Genetics 12: 1241-1252.
Beekmans S and Van Driessche E. Pig heart fumarase contains two distinct substrate-binding sites
differing in affinity. (1998) Journal of Biological Chemistry 273(48):
31661-31669.
Despoisses S, Noel L, Choiset A, Portnoi MF, Turleau C,
Quack B, Taillemite JL, de Grouchy J, Junien C. (1984) Regional mapping of FH to band 1q42.1 by gene dosage studies.
Cytogenetics and Cell Genetics 37: 450-451.
Massey V. (1953) Studies on fumarase. 4. The effects of
inhibitors on fumarase activity. Biochemical Journal 55(1): 172–177.
Weaver, T.M., Levitt, D.G., Donnelly, M.I., Stevens, P.P. and Banaszak, L.J. (1995) The multisubunit active site of fumarase C from Escherichia coli. Nature Structural Biology 2: 654–662.
Weaver, T. and Banaszak, L. (1996) Crystallographic studies of the catalytic and a second site in fumarase C from Escherichia coli. Biochemistry 35: 13955–13965.
Diagram of the reaction catalyzed by the
enzyme, all reactants and coenzymes, free energy change if available, any known
inhibitors, activators, responses
to hormone signals. Major metabolic role. Structural
information about enzyme: number of subunits, shape and size,
x ray crystallography and/or NMR structures, active site shape/size. If possible, get
the 3D image of the protein from SwissProt and include it, crediting
source.